Python for bioinformatics
Marcello Beltrami
Bioinformatician

Bioinformatics is a discipline combining the use of computational techniques to extract biological insights from diverse biological data. These include biological fields such as metabolomics, genomic sequencing, and proteomics. A new field that has recently seen a higher research output is system biology, which involves the use of multiple data sources to model how a biological system works. Example of these can be seen throughout literature involving water fleas (*Daphnia pulex)* as a model organism used to monitor early effects of pollution in the environment. As computers are effectively fancy rocks with electricity flowing through them, to effectively analyse all data generated we need to tell them how to automate all the algorithmic, statistical, and mathematical calculations required to extract valuable biological insights. In turn, this requires an extensive knowledge of the computational field, and software development practices. This further translates in choosing what for the computer scientist is the right chisel to the sculptor: a programming language.
Python in Bioinformatics
For many, python is the introductory language to programming in general. Although this has been said countless times, python reads like English, making grasping computational easier than other languages. This creates a vast community of passionate developers which create libraries for pretty much any purpose.
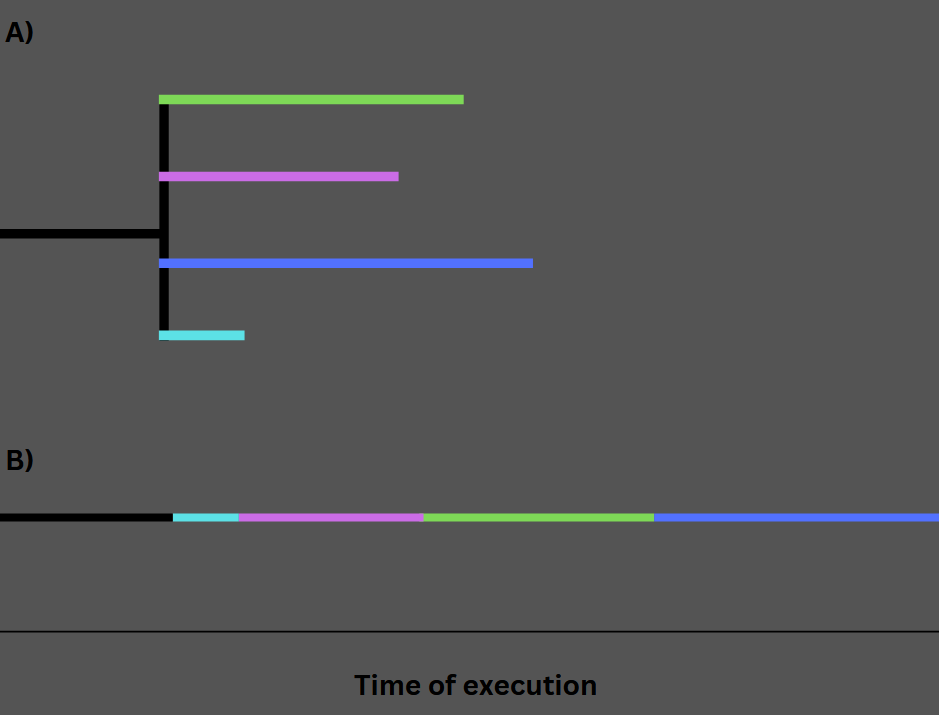
Performance and package management
The presence of the pip package manager, combined with environment such Anaconda allow to create a relatively streamline process when it comes to development. The best thing about Python is the highly modular system and extensive library availability. Also, generally speaking libraries syntax is standardised, which is definite pro when compared to other languages such R. Examples of how bioinformatics tooling have been published through pip can be seen as CNVKit and BlastWrapper (shameless plug I know). This allows to manage dependencies, further ensuring reproducibility. Python’s performance is the major issue due to the GIL (Global Interpreter Lock). The GIL is a mutual exclusion lock used by CPython to ensure that only one thread executes Python bytecode at a time, ensuring garbage collection and memory safety. This results in lack of true multithreading in the language, which further contribute to slower data processing when compared to other lower level languages. Despite this, there are some workarounds the lack of native multithreading. Examples involve the use of computational paradigms such as async programming, which leverages asynchronous processing. This process involves executing a task via an API over the internet or as a local subprocess (e.g., Samtools) and awaiting the response upon completion. Since each data entry can be processed independently, this approach significantly enhances performance when interacting with APIs. Without parallelization, the delay in receiving query results could considerably slow down the tool.

Why some software developers dislike python
Some developers have previously argued that, although Python’s readability is unparallel, using it to develop data processing tools is an overall poor choice. As previously covered, once the ability of a developer is removed from the equation, Python is really slow when compared to other available languages. Additionally, the low barrier of entry acts as a double edge sword, as this results in programmers with poor knowledge of CompSci concepts such as algorithms, data structures and hardware knowledge to have access to programming, resulting in suboptimal software implementations. Granted, this is not a rant against those programmer (I am one of them LOL), but it is something that should be acknowledged when choosing how to tackle a problem, further supporting the thesis that bioinformatics is a widely collaborative field, and that gatekeeping can only harm research. At the end of the day, language is just a tool to interface yourself with a computer. In fact, Python if often better reserved as a glue language, where the heavy backend computations is carried by libraries written in C, C++ or dare I say Rust.
Pipeline development using python in bioinformatics.
So, the best applications for python are reserved to glue scripts which used pre-compiled lower level libraries or using it for data analysis via the beautiful integration in Jupyter Notebook, which combined with environment creation allows to create highly reproducible pipelines. Some project ideas could be the use of standard Linux tooling such as grep, awk and etc to parse common files formats such as BAM, VCF or BED to extract insights. Quite commonly, good chunk of bioinformatics revolves around parsing files in search for specific patterns.
Marcello Beltrami
Bioinformatician